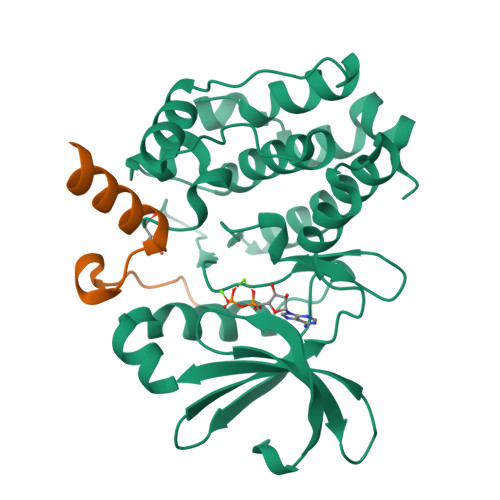
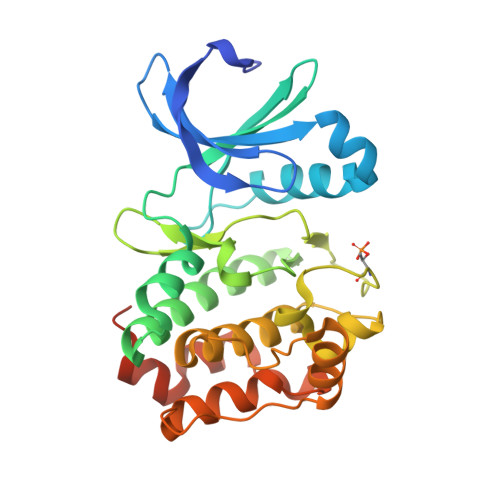
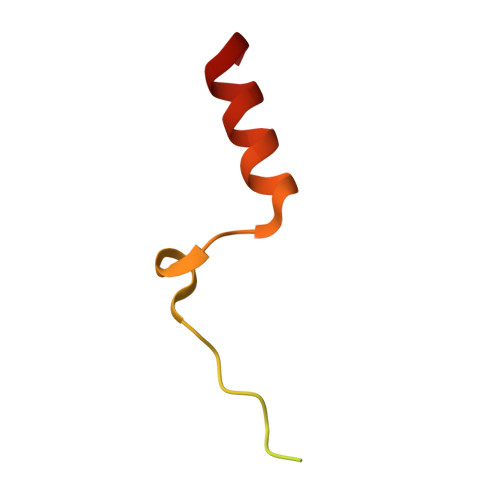
Structural Basis of N-Myc Binding by Aurora-A and its Destabilization by Kinase Inhibitors
Richards, M., Burgess, S., Poon, E., Carstensen, A., Eilers, M., Chesler, L., Bayliss, R.(2016) Proc Natl Acad Sci U S A 113: 13726
- PubMed: 27837025
- DOI: https://doi.org/10.1073/pnas.1610626113
- Primary Citation of Related Structures:
5G1X - PubMed Abstract:
Myc family proteins promote cancer by inducing widespread changes in gene expression. Their rapid turnover by the ubiquitin-proteasome pathway is regulated through phosphorylation of Myc Box I and ubiquitination by the E3 ubiquitin ligase SCF FbxW7 However, N-Myc protein (the product of the MYCN oncogene) is stabilized in neuroblastoma by the protein kinase Aurora-A in a manner that is sensitive to certain Aurora-A-selective inhibitors. Here we identify a direct interaction between the catalytic domain of Aurora-A and a site flanking Myc Box I that also binds SCF FbxW7 We determined the crystal structure of the complex between Aurora-A and this region of N-Myc to 1.72-Å resolution. The structure indicates that the conformation of Aurora-A induced by compounds such as alisertib and CD532 is not compatible with the binding of N-Myc, explaining the activity of these compounds in neuroblastoma cells and providing a rational basis for the design of cancer therapeutics optimized for destabilization of the complex. We also propose a model for the stabilization mechanism in which binding to Aurora-A alters how N-Myc interacts with SCF FbxW7 to disfavor the generation of Lys48-linked polyubiquitin chains.
Organizational Affiliation:
Astbury Centre for Structural and Molecular Biology, Faculty of Biological Sciences, University of Leeds, Leeds LS2 9JT, United Kingdom.